Machine learning pipeline for detection of boundaries around Mercury
Inhalt
4. Machine learning pipeline for detection of boundaries around Mercury¶
4.1. Installation¶
Step 1: Download the data from http://epn2024.sinp.msu.ru/epsc2021/ and save it under data/
Step 2: Install the conda environmet : conda env create -f environment.yml
Step 3: Run jupyter notebook Magnetometer.ipynb
4.2. Imports¶
### standard imports
from __future__ import division
import numpy as np
import pandas as pd
import os, copy
import re
import pickle
import time
### sklearn imports
from sklearn import metrics
### pytorch imports
import torch
import torch.nn as nn
from torch.nn.parameter import Parameter
import torch.nn.functional as F
import torchvision.transforms as transforms
import torch.utils.data as data
from torch.autograd import Variable
torch.manual_seed(0)
## plotting imports
import matplotlib.pyplot as plt
import seaborn as sns
import plotly.graph_objs as go
from IPython.display import Image
### silence future warnings
import warnings
warnings.simplefilter(action='ignore', category=FutureWarning)
---------------------------------------------------------------------------
ModuleNotFoundError Traceback (most recent call last)
/var/folders/x3/2bzh843n0tv469w6l6sd8sq00000gn/T/ipykernel_23905/2753096250.py in <module>
12
13 ### pytorch imports
---> 14 import torch
15 import torch.nn as nn
16 from torch.nn.parameter import Parameter
ModuleNotFoundError: No module named 'torch'
4.3. Example orbit data¶
df = pd.read_csv('../../../../labelled_orbits/train/df_10.csv')
df.head()
| Unnamed: 0 | DATE | X_MSO | Y_MSO | Z_MSO | BX_MSO | BY_MSO | BZ_MSO | DBX_MSO | DBY_MSO | ... | Z | VX | VY | VZ | VABS | D | COSALPHA | EXTREMA | ORBIT | LABEL | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 347717 | 2011-03-28 08:17:09 | 8216.028 | 2915.743 | -15375.351 | 27.674 | -11.254 | 12.281 | 0.136 | 0.316 | ... | 6.238218e+06 | -36.116236 | -39.06727 | 0.121972 | 53.203843 | 5.181665e+07 | 0.873091 | 2 | 10 | 0 |
| 1 | 347718 | 2011-03-28 08:17:10 | 8216.439 | 2916.030 | -15375.077 | 26.448 | -11.211 | 13.221 | 0.515 | 0.338 | ... | 6.238218e+06 | -36.116236 | -39.06727 | 0.121909 | 53.203843 | 5.181665e+07 | 0.873091 | 0 | 10 | 0 |
| 2 | 347719 | 2011-03-28 08:17:11 | 8216.850 | 2916.316 | -15374.803 | 26.505 | -11.706 | 12.536 | 0.251 | 1.426 | ... | 6.238218e+06 | -36.116236 | -39.06727 | 0.121926 | 53.203843 | 5.181665e+07 | 0.873091 | 0 | 10 | 0 |
| 3 | 347720 | 2011-03-28 08:17:12 | 8217.261 | 2916.603 | -15374.529 | 25.981 | -18.053 | 6.531 | 0.379 | 1.536 | ... | 6.238218e+06 | -36.116236 | -39.06727 | 0.121968 | 53.203843 | 5.181665e+07 | 0.873091 | 0 | 10 | 0 |
| 4 | 347721 | 2011-03-28 08:17:13 | 8217.671 | 2916.889 | -15374.255 | 25.847 | -18.826 | 5.695 | 0.103 | 0.301 | ... | 6.238218e+06 | -36.116236 | -39.06727 | 0.121897 | 53.203843 | 5.181665e+07 | 0.873091 | 0 | 10 | 0 |
5 rows × 32 columns
df.columns
Index(['Unnamed: 0', 'DATE', 'X_MSO', 'Y_MSO', 'Z_MSO', 'BX_MSO', 'BY_MSO',
'BZ_MSO', 'DBX_MSO', 'DBY_MSO', 'DBZ_MSO', 'RHO_DIPOLE', 'PHI_DIPOLE',
'THETA_DIPOLE', 'BABS_DIPOLE', 'BX_DIPOLE', 'BY_DIPOLE', 'BZ_DIPOLE',
'RHO', 'RXY', 'X', 'Y', 'Z', 'VX', 'VY', 'VZ', 'VABS', 'D', 'COSALPHA',
'EXTREMA', 'ORBIT', 'LABEL'],
dtype='object')
df[['BX_MSO', 'BY_MSO', 'BZ_MSO']].plot(figsize = (13,8))
<AxesSubplot:>
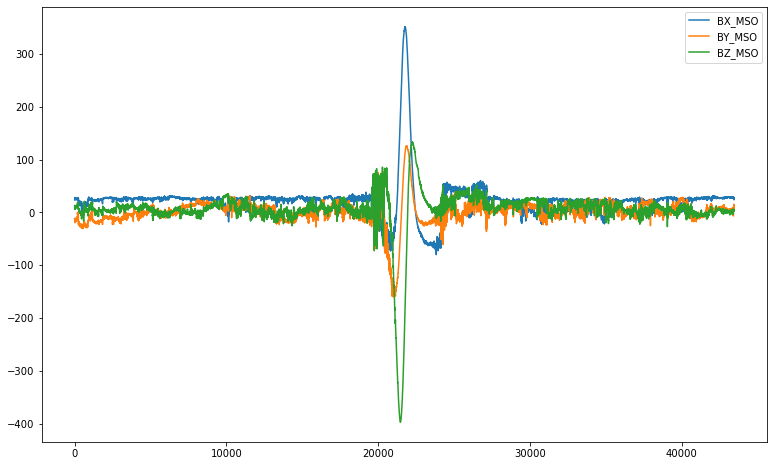
4.4. Data Description¶
X_MSO : Spacecraft position X in MSO coord system [km]
Y_MSO : Spacecraft position Y in MSO coord system [km]
Z_MSO : Spacecraft position Z in MSO coord system [km]
BX_MSO: Magnetic field X component in MSO coord system [nT]
BY_MSO: Magnetic field Y component in MSO coord system [nT]
BZ_MSO: Magnetic field X component in MSO coord system [nT]
DBX_MSO: Magnetic field X component in MSO coord system [nT]
DXY_MSO: Magnetic field Y component in MSO coord system [nT]
DBZ_MSO: Magnetic field X component in MSO coord system [nT]
RHO: Planetocentric spacecraft distance [km]
RXY: Spacecraft distance to the MSO Z axis [km]
RHO_DIPOLE : Dipole-centric spacecraft distance [km]
THETA_DIPOLE: Spacecraft magnetic latitude [rad]
BABS_DIPOLE: Planetary dipole total magnetic field magnitude [nT]
BX_DIPOLE: Planetary dipole magnetic field XMSO component [nT]
BY_DIPOLE: Planetary dipole magnetic field in YMSO component [nT]
BZ_DIPOLE: Planetary dipole magnetic field ZMSO componenet [nT]
X : Mercury Heliocentric position X in SE coordinate system [km]
Y : Mercury Heliocentric position Y in SE coordinate system [km]
Z : Mercury Heliocentric position Z in SE coordinate system [km]
VX : Mercury orbital velocity component in SE coord system [km/sec]
VY : Mercury orbital velocity component in SE coord system [km/sec]
VZ : Mercury orbital velocity component in SE coord system [km/sec]
VABS: Mercury orbital velocity magnitude [km/sec]
D: Mercuty heliocentric distance [km]
COSALPHA: Cosine of Mercury azimuth angle in SE coord system
EXTREMA: 1 in apoapsis, -1 in periapsis, 0 otherwise
### important column names
DATE_COL = "DATE"
ORBIT_COL = "ORBIT"
LABEL_COL = "LABEL"
### relevant feature column names
COLS_3D = [
["X", "Y", "Z"],
["VX", "VY", "VZ"],
["X_MSO", "Y_MSO", "Z_MSO"],
["BX_MSO", "BY_MSO", "BZ_MSO"],
["DBX_MSO", "DBY_MSO", "DBZ_MSO"],
["BX_DIPOLE", "BY_DIPOLE", "BZ_DIPOLE"],
]
COLS_SINGLE = [
"RHO_DIPOLE",
"PHI_DIPOLE",
"THETA_DIPOLE",
"BABS_DIPOLE",
"RHO", "RXY",
"VABS", "D",
"COSALPHA"
]
FLUX_COLS = COLS_3D[3]
### class name mapping
LABEL_NAMES = {
0: "interplanetary magnetic field",
1: "bow shock crossing",
2: "magnetosheath",
3: "magnetopause crossing",
4: "magnetosphere"
}
### event columns in label file
EVENT_COLS = list(range(1, 9))
### Helper functions to normalise data
def normalize_decoupled(data, cols):
data[cols] = (data[cols] - data[cols].mean()) / data[cols].std()
def normalize_coupled(data, cols):
centered = data[cols] - data[cols].mean()
distdev = (centered ** 2).sum(axis=1).mean() ** 0.5
data[cols] = centered / distdev
4.5. Prepare Dataloader¶
4.5.1. (a) with sliding window¶
### Data loader with sliding window
class MessengerDataset(data.Dataset):
"""
The MESSENGER dataset, cut into fixed-length time windows.
"""
# TODO add parameters for 3D and 1D feature column names
def __init__(self, orbits_path, *, window_size=8, normalize=True):
"""
Loads the MESSENGER dataset from orbits_path and configures it according to the parameters.
Parameters
----------
orbits_path : str
The path to the folder containing all MESSENGER orbit data.
window_size : int, default 8
The size each window shall have.
normalize : bool, default True
Whether to normalize the features.
"""
self.data = pd.read_csv(orbits_path)
self.window_size = window_size
self.skip_size = window_size - 1
orbit_sizes = self.data.groupby(ORBIT_COL).size()
self.cum_window_nums = (orbit_sizes - self.skip_size).cumsum()
if normalize:
for feat_3d in COLS_3D:
normalize_coupled(self.data, feat_3d)
normalize_decoupled(self.data, COLS_SINGLE)
def __len__(self):
"""
Yields the size of the dataset.
Returns
-------
int
The total number of windows.
"""
return self.cum_window_nums.iloc[-1]
def __getitem__(self, idx):
"""
Retrieves the window the given index and its corresponding label.
Parameters
----------
idx
The index of the desired window.
Returns
-------
sample : bool
The requested window as float tensor.
label : int
The corresponding label between 0 and 4.
Raises
------
IndexError
If idx is negative or larger than the dataset size.
"""
if idx < 0 or idx >= len(self):
raise IndexError(f"Index {idx} is out of bounds.")
orbit_skips = (self.cum_window_nums <= idx).sum()
pos = idx + orbit_skips * self.skip_size
window = self.data.iloc[pos:(pos + self.window_size)]
feats_3d = [torch.tensor(window[feat].values) for feat in COLS_3D]
sample = torch.cat(feats_3d).view(self.window_size, -1, 3) # window_size x #feats_3d x 3
label = torch.tensor(window[LABEL_COL].values) # list of ordinal labels
return sample, label
dataset = Magnetometer_data('../../../../labelled_orbits/train/df_10.csv', features)
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
<ipython-input-57-cfdba41dad21> in <module>
----> 1 dataset = Magnetometer_data('../../../../labelled_orbits/train/df_10.csv', features)
NameError: name 'features' is not defined
len(dataset)
43466
4.5.2. (b) without sliding window¶
### Data loader without windowing
class Magnetometer_data(data.Dataset):
def __init__(self, csv_file , features, sample_rate = 1, transform = None, root = ''):
"""Args:
root (string): Base path
features (list): selected features
csv_file (string): Path to csv file with sensor data
transform (callable, optional): Transforms to be applied.
"""
assert len(features) % 3 == 0
self.data_path = os.path.join(root, csv_file)
self.feature_cols = features
self.sample_rate = sample_rate
self.data = pd.read_csv(self.data_path, usecols = self.feature_cols).dropna()
# self.data = self.__normalise__(self.data)
self.transform = transform
try:
self.labels = pd.read_csv(self.data_path, usecols = ['LABEL'])
except ValueError as e:
self.labels = None
def __len__(self):
return len(self.data)
def __label_to_int(self, label):
label_ord = {'IMF': 0, 'BS-crossing': 1, 'MP-crossing' : 3, 'magnetosheath': 2, 'magnetosphere': 4}
return label_ord[label]
def __normalise__(self, features):
mean = np.mean(features)
std = np.std(features)
return (features - mean)/std
def __reshape__(self, x):
##reshape bands, features
return x.reshape(-1,3)
#return torch.unsqueeze(x,0)
def __getitem__(self, idx):
coords = self.__reshape__(torch.tensor(self.data.iloc[idx].values))
if self.transform:
coords = self.transform(coords)
if self.labels is not None:
#label_int = torch.tensor(self.__label_to_int(self.labels.iloc[idx]['labels']))
label_int = self.labels.iloc[idx]['LABEL']
sample = {'coords': coords, 'labels': label_int}
return sample
else:
sample = {'coords': coords}
return sample
4.6. Convolutions¶
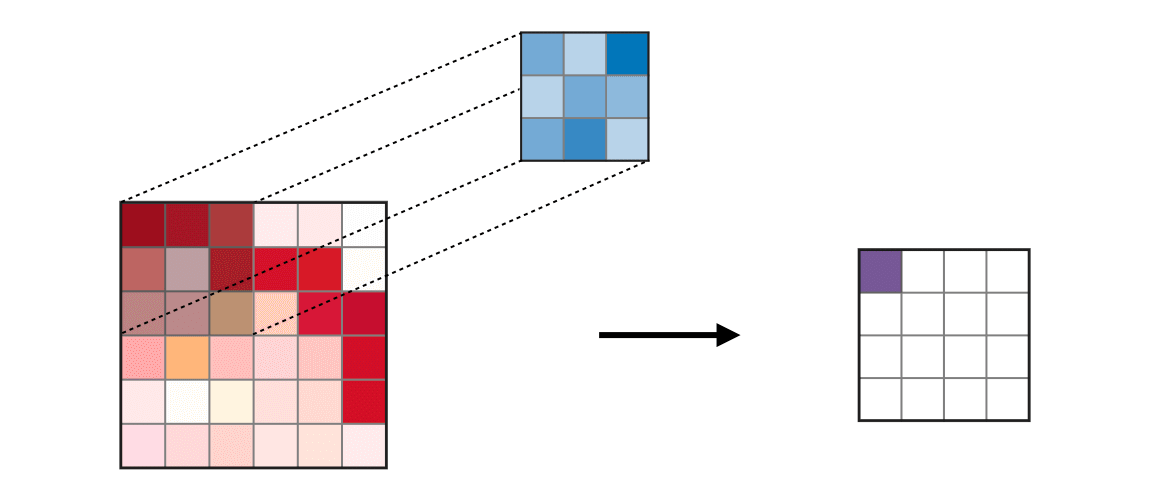
The convolution layer uses filters that perform convolution operations as it is scanning the input \(I\) with respect to its dimensions. Its hyperparameters include the filter size \(F\) and stride \(S\). The resulting output \(O\) is called feature map or activation map.
Image('img/convolution-layer-a.png')
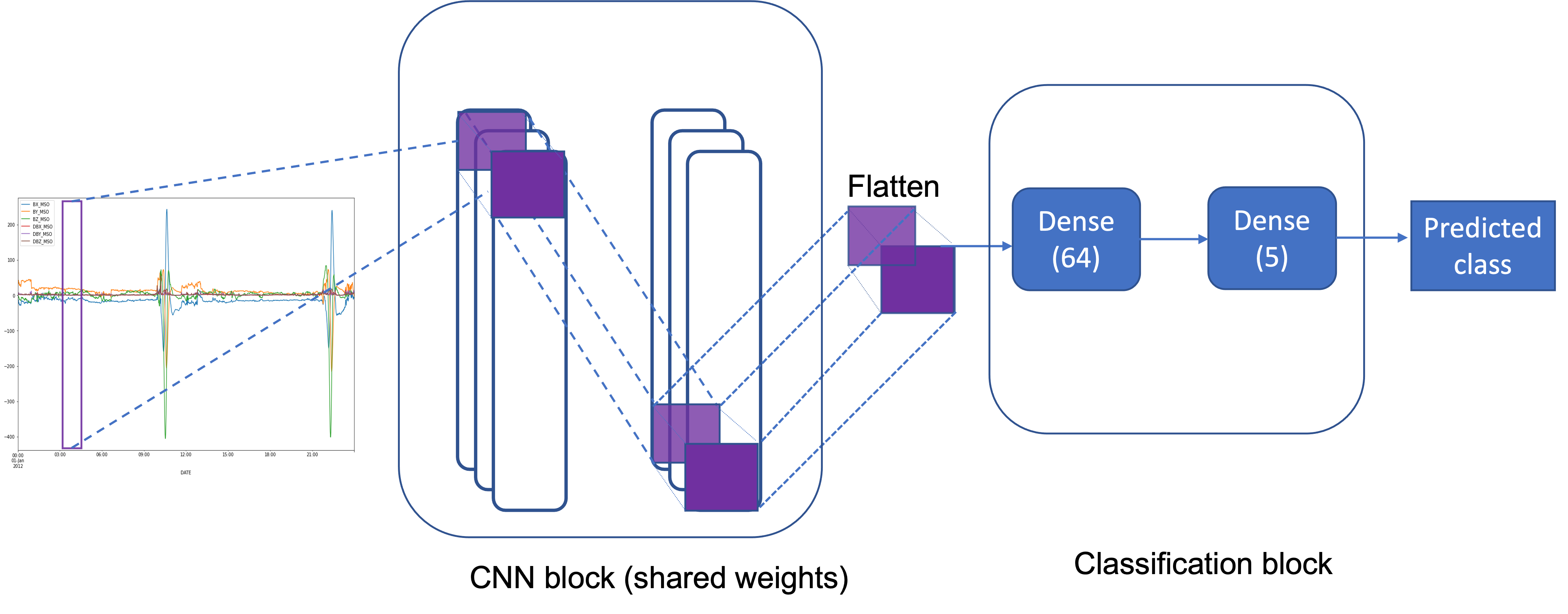
class CNN(nn.Module):
def __init__(self, input_dim = 3, bands = 1):
"""
Description:
3 layer simple CNN followed by 2 FC layers.
Args:
input_dim: dimensions in the data, default = 3 for X, Y,Z
bands : number of features/channels. eg: Aircraft position --> 1
"""
super(CNN, self).__init__()
self.bands = bands
self.conv1 = nn.Conv1d(input_dim, 32, 1)
self.conv2 = nn.Conv1d(32, 64, 1)
self.conv3 = nn.Conv1d(64, 128, 1)
self.fc1 = nn.Linear(128 * self.bands , 64)
self.fc2 = nn.Linear(64, 5)
self.activation = nn.LogSoftmax(dim = 1)
def forward(self, x):
x = F.relu(self.conv1(x))
x = F.relu(self.conv2(x))
x = F.relu(self.conv3(x))
x = x.reshape(-1, 128 * self.bands)
x = F.relu(self.fc1(x))
x = self.activation(self.fc2(x))
return x
Image('img/cnn_magnetometer.png')
### Simple n layer LSTM network, modified from nn.LSTM
class LSTM(nn.Module):
def __init__(self, input_dim, hidden_dim, layer_dim, output_dim):
"""
Description:
Simple LSTM with recurrence
Args:
input_dim: dimensions in the data, default
layer_dim: number of layers
output_dim: output units
"""
super(LSTM, self).__init__()
# Hidden dimensions
self.hidden_dim = hidden_dim
# Number of hidden layers
self.layer_dim = layer_dim
# batch_first=True causes input/output tensors to be of shape
# (batch_dim, seq_dim, feature_dim)
self.lstm = nn.LSTM(input_dim, hidden_dim, layer_dim, batch_first=True)
# Readout layer
self.fc = nn.Linear(hidden_dim*2, output_dim)
def forward(self, x):
b = x.size(0)
# Initialize hidden state with zeros
h0 = torch.zeros(self.layer_dim, x.size(0), self.hidden_dim, device = device,).requires_grad_()
# Initialize cell state
c0 = torch.zeros(self.layer_dim, x.size(0), self.hidden_dim, device = device).requires_grad_()
# We need to detach for backpropagation through time (BPTT)
# If we don't, we'll backprop all the way to the start even after going through another batch
out, (hn, cn) = self.lstm(x, (h0.detach(), c0.detach()))
# Index hidden state of last time step
out = self.fc(torch.reshape(out,(b,-1)))
return out
class CRNN(nn.Module):
def __init__(self, input_dim = 16, hidden_dim = 16, n_layers = 2,bands = 1):
"""
Args:
input_dim: dimensions in the data, default = 3 for X, Y,Z
bands : number of features/channels. eg: Aircraft position --> 1
"""
super(CRNN, self).__init__()
self.bands = bands
self.hidden_dim = hidden_dim
self.n_layers = n_layers
self.cnn1 = nn.Conv1d(input_dim, 32,1)
self.cnn2 = nn.Conv1d(32, 64, 1 )
self.bn1 = nn.BatchNorm1d(32)
self.bn2 = nn.BatchNorm1d(64)
self.fc0 = nn.Linear(hidden_dim, 8)
self.fc1 = nn.Linear(8 , 5)
#self.fc4 = nn.Linear(64, 5)
self.activation = nn.LogSoftmax(dim = -1)
self.rnn = nn.RNN(64, self.hidden_dim, self.n_layers, batch_first=True)
def init_hidden(self, batch_size):
# This method generates the first hidden state of zeros which we'll use in the forward pass
# We'll send the tensor holding the hidden state to the device we specified earlier as well
hidden = torch.zeros(self.n_layers, batch_size, self.hidden_dim).to(device)
return hidden
def forward(self, x):
b = x.size(0)
t = x.size(2)
#h,w = x.size(2), x.size(3)
x = F.relu(self.bn1(self.cnn1(x)))
x = F.relu(self.bn2(self.cnn2(x)))
x = x.transpose(1,2)
# Initializing hidden state for first input using method defined below
hidden = self.init_hidden(b)
# Passing in the input and hidden state into the model and obtaining outputs
out, hidden = self.rnn(x, hidden)
hidden = hidden.permute(0,2,1)
#hidden = hidden.view(b,t,-1)
out = out.contiguous().view(b,t,-1)
x = F.relu((self.fc0(out)))
x = F.relu((self.fc1(x)))
x = self.activation(x)
#x = x.reshape(b, t, 5)
return x
class CNN_window(nn.Module):
def __init__(self, input_dim = 3, bands = 1):
"""
Args:
input_dim: dimensions in the data, default = 3 for X, Y,Z
bands : number of features/channels. eg: Aircraft position --> 1
"""
super(CNN_window, self).__init__()
self.bands = bands
self.conv1 = nn.Conv1d(input_dim, 32, 1)
self.bn1 = nn.BatchNorm1d(32)
self.conv2 = nn.Conv1d(32, 64, 1)
self.bn2 = nn.BatchNorm1d(64)
self.conv3 = nn.Conv1d(64, 128, 1)
self.bn3 = nn.BatchNorm1d(128)
self.fc0 = nn.Linear(128,64)
self.fc1 = nn.Linear(1, 8)
self.fc2 = nn.Linear(64 , 32)
self.fc3 = nn.Linear(32, 5)
#self.fc4 = nn.Linear(64, 5)
self.activation = nn.Softmax(dim = -1)
def forward(self, x):
x = F.relu(self.bn1(self.conv1(x)))
x = F.relu(self.bn2(self.conv2(x)))
x = F.relu(self.bn3(self.conv3(x)))
x = x.transpose(1,2)
x = F.relu(self.fc0(x))
activations = F.relu(self.fc2(x))
output = self.activation(self.fc3(activations))
return output
def train(model, optimizer , train_loader, val_loader,criterion = nn.CrossEntropyLoss(),save = False, epochs = 1):
# iteration_loss = []
for epoch in range(epochs):
model.train()
running_loss = 0.0
for i,sample in enumerate(train_loader,0):
## load inputs and labels
inputs = sample[0].float().to(device)
inputs = inputs.squeeze(1)
b = inputs.size(0)
t = inputs.size(1)
inputs = inputs.reshape(b, t, -1)
labels = sample[1].long().to(device)
labels = labels.squeeze(1)
# labels = torch.nn.functional.one_hot(labels_int)
## set grad to 0
optimizer.zero_grad()
## forward propagate
output = model(inputs.transpose(1,2))
## Calculate classification output
_, predicted = torch.max(output, -1)
loss = criterion(output.view(-1,5), labels.view(-1))
#loss = criterion(output, labels)
## backward + optimize only if in training phase
loss.backward()
## update the weights
optimizer.step()
## print statistics
running_loss += loss.item()
if i % 10 == 9: # print every 10 mini-batches
print('[Epoch: %d, Batch: %4d / %4d], loss: %.3f' %
(epoch + 1, i + 1, len(train_loader), running_loss / 10))
running_loss = 0.0
# iteration_loss.append(running_loss)
## validation
if val_loader:
model.eval()
correct = total = 0
with torch.no_grad():
for sample in val_loader:
inputs = sample[0].float().to(device)
inputs = inputs.squeeze(1)
b = inputs.size(0)
t = inputs.size(1)
inputs = inputs.reshape(b, t, -1)
labels = sample[1].long().to(device)
labels = labels.squeeze(1)
output = model(inputs.transpose(1,2))
_, predicted = torch.max(output, -1)
total += labels.size(0)
correct += (predicted == labels).sum().item()
val_acc = 100. * correct / total
print('Valid accuracy: %d %%' % val_acc)
## save
if save:
torch.save(model.state_dict(), "save_"+str(epoch)+".pth")
# Load device
device = torch.device("cuda:0" if torch.cuda.is_available() else "cpu")
# Parameters
params = {'batch_size': 4096,
'shuffle': True,
'num_workers': 32}
csv = 'data/sampled_df/df_train.csv'
csv_test = 'data/sampled_df/df_test.csv'
xdata = MessengerDataset(csv, window_size = 1)
test_data = MessengerDataset(csv_test, window_size = 1)
# Train, validation split
# 80/20 split
#train_set, val_set = xdata[: 80], xdata[int(len(xdata)*0.8):]
#train_set, val_set = torch.utils.data.random_split(xdata, [int(len(xdata)*0.8), int(len(xdata)*0.2)])
### Create dataloader
train_loader = torch.utils.data.DataLoader(xdata, **params)
validation_loader = torch.utils.data.DataLoader(test_data, **params)
### Load model
model = CNN(3,6).to(device)
optimizer = torch.optim.Adam(model.parameters(), lr = 0.0001)
train(model, optimizer, train_loader, validation_loader, epochs = 150, save = False)
[Epoch: 1, Batch: 10 / 3849], loss: 1.589
[Epoch: 1, Batch: 20 / 3849], loss: 1.540
[Epoch: 1, Batch: 30 / 3849], loss: 1.484
[Epoch: 1, Batch: 40 / 3849], loss: 1.419
[Epoch: 1, Batch: 50 / 3849], loss: 1.344
[Epoch: 1, Batch: 60 / 3849], loss: 1.263
[Epoch: 1, Batch: 70 / 3849], loss: 1.175
[Epoch: 1, Batch: 80 / 3849], loss: 1.096
[Epoch: 1, Batch: 90 / 3849], loss: 1.020
[Epoch: 1, Batch: 100 / 3849], loss: 0.963
[Epoch: 1, Batch: 110 / 3849], loss: 0.922
[Epoch: 1, Batch: 120 / 3849], loss: 0.887
[Epoch: 1, Batch: 130 / 3849], loss: 0.860
[Epoch: 1, Batch: 140 / 3849], loss: 0.824
[Epoch: 1, Batch: 150 / 3849], loss: 0.795
[Epoch: 1, Batch: 160 / 3849], loss: 0.761
[Epoch: 1, Batch: 170 / 3849], loss: 0.742
[Epoch: 1, Batch: 180 / 3849], loss: 0.709
[Epoch: 1, Batch: 190 / 3849], loss: 0.677
[Epoch: 1, Batch: 200 / 3849], loss: 0.650
[Epoch: 1, Batch: 210 / 3849], loss: 0.622
[Epoch: 1, Batch: 220 / 3849], loss: 0.593
[Epoch: 1, Batch: 230 / 3849], loss: 0.562
[Epoch: 1, Batch: 240 / 3849], loss: 0.537
[Epoch: 1, Batch: 250 / 3849], loss: 0.509
[Epoch: 1, Batch: 260 / 3849], loss: 0.496
[Epoch: 1, Batch: 270 / 3849], loss: 0.468
[Epoch: 1, Batch: 280 / 3849], loss: 0.462
[Epoch: 1, Batch: 290 / 3849], loss: 0.435
[Epoch: 1, Batch: 300 / 3849], loss: 0.424
[Epoch: 1, Batch: 310 / 3849], loss: 0.409
[Epoch: 1, Batch: 320 / 3849], loss: 0.394
[Epoch: 1, Batch: 330 / 3849], loss: 0.382
[Epoch: 1, Batch: 340 / 3849], loss: 0.378
[Epoch: 1, Batch: 350 / 3849], loss: 0.366
[Epoch: 1, Batch: 360 / 3849], loss: 0.352
[Epoch: 1, Batch: 370 / 3849], loss: 0.339
[Epoch: 1, Batch: 380 / 3849], loss: 0.337
[Epoch: 1, Batch: 390 / 3849], loss: 0.322
[Epoch: 1, Batch: 400 / 3849], loss: 0.323
[Epoch: 1, Batch: 410 / 3849], loss: 0.313
[Epoch: 1, Batch: 420 / 3849], loss: 0.311
[Epoch: 1, Batch: 430 / 3849], loss: 0.305
[Epoch: 1, Batch: 440 / 3849], loss: 0.297
[Epoch: 1, Batch: 450 / 3849], loss: 0.296
[Epoch: 1, Batch: 460 / 3849], loss: 0.287
[Epoch: 1, Batch: 470 / 3849], loss: 0.278
[Epoch: 1, Batch: 480 / 3849], loss: 0.283
[Epoch: 1, Batch: 490 / 3849], loss: 0.278
[Epoch: 1, Batch: 500 / 3849], loss: 0.274
[Epoch: 1, Batch: 510 / 3849], loss: 0.267
[Epoch: 1, Batch: 520 / 3849], loss: 0.265
[Epoch: 1, Batch: 530 / 3849], loss: 0.263
[Epoch: 1, Batch: 540 / 3849], loss: 0.258
[Epoch: 1, Batch: 550 / 3849], loss: 0.256
[Epoch: 1, Batch: 560 / 3849], loss: 0.258
[Epoch: 1, Batch: 570 / 3849], loss: 0.255
[Epoch: 1, Batch: 580 / 3849], loss: 0.248
[Epoch: 1, Batch: 590 / 3849], loss: 0.251
[Epoch: 1, Batch: 600 / 3849], loss: 0.245
[Epoch: 1, Batch: 610 / 3849], loss: 0.247
[Epoch: 1, Batch: 620 / 3849], loss: 0.246
[Epoch: 1, Batch: 630 / 3849], loss: 0.238
[Epoch: 1, Batch: 640 / 3849], loss: 0.244
[Epoch: 1, Batch: 650 / 3849], loss: 0.232
[Epoch: 1, Batch: 660 / 3849], loss: 0.238
[Epoch: 1, Batch: 670 / 3849], loss: 0.238
[Epoch: 1, Batch: 680 / 3849], loss: 0.232
[Epoch: 1, Batch: 690 / 3849], loss: 0.239
[Epoch: 1, Batch: 700 / 3849], loss: 0.230
[Epoch: 1, Batch: 710 / 3849], loss: 0.232
[Epoch: 1, Batch: 720 / 3849], loss: 0.234
[Epoch: 1, Batch: 730 / 3849], loss: 0.228
[Epoch: 1, Batch: 740 / 3849], loss: 0.226
[Epoch: 1, Batch: 750 / 3849], loss: 0.227
[Epoch: 1, Batch: 760 / 3849], loss: 0.229
[Epoch: 1, Batch: 770 / 3849], loss: 0.221
[Epoch: 1, Batch: 780 / 3849], loss: 0.219
[Epoch: 1, Batch: 790 / 3849], loss: 0.219
[Epoch: 1, Batch: 800 / 3849], loss: 0.222
[Epoch: 1, Batch: 810 / 3849], loss: 0.221
[Epoch: 1, Batch: 820 / 3849], loss: 0.217
[Epoch: 1, Batch: 830 / 3849], loss: 0.215
[Epoch: 1, Batch: 840 / 3849], loss: 0.223
[Epoch: 1, Batch: 850 / 3849], loss: 0.215
[Epoch: 1, Batch: 860 / 3849], loss: 0.219
[Epoch: 1, Batch: 870 / 3849], loss: 0.218
[Epoch: 1, Batch: 880 / 3849], loss: 0.215
[Epoch: 1, Batch: 890 / 3849], loss: 0.214
[Epoch: 1, Batch: 900 / 3849], loss: 0.218
[Epoch: 1, Batch: 910 / 3849], loss: 0.210
[Epoch: 1, Batch: 920 / 3849], loss: 0.213
[Epoch: 1, Batch: 930 / 3849], loss: 0.206
[Epoch: 1, Batch: 940 / 3849], loss: 0.213
[Epoch: 1, Batch: 950 / 3849], loss: 0.209
[Epoch: 1, Batch: 960 / 3849], loss: 0.208
[Epoch: 1, Batch: 970 / 3849], loss: 0.206
[Epoch: 1, Batch: 980 / 3849], loss: 0.205
[Epoch: 1, Batch: 990 / 3849], loss: 0.204
[Epoch: 1, Batch: 1000 / 3849], loss: 0.208
[Epoch: 1, Batch: 1010 / 3849], loss: 0.205
[Epoch: 1, Batch: 1020 / 3849], loss: 0.203
[Epoch: 1, Batch: 1030 / 3849], loss: 0.200
[Epoch: 1, Batch: 1040 / 3849], loss: 0.200
[Epoch: 1, Batch: 1050 / 3849], loss: 0.204
[Epoch: 1, Batch: 1060 / 3849], loss: 0.202
[Epoch: 1, Batch: 1070 / 3849], loss: 0.198
[Epoch: 1, Batch: 1080 / 3849], loss: 0.202
---------------------------------------------------------------------------
KeyboardInterrupt Traceback (most recent call last)
<ipython-input-58-e70daca3e090> in <module>
2 model = CNN(3,6).to(device)
3 optimizer = torch.optim.Adam(model.parameters(), lr = 0.0001)
----> 4 train(model, optimizer, train_loader, validation_loader, epochs = 150, save = False)
<ipython-input-55-272714bc5674> in train(model, optimizer, train_loader, val_loader, criterion, save, epochs)
4 model.train()
5 running_loss = 0.0
----> 6 for i,sample in enumerate(train_loader,0):
7 ## load inputs and labels
8 inputs = sample[0].float().to(device)
~/anaconda3/lib/python3.8/site-packages/torch/utils/data/dataloader.py in __next__(self)
515 if self._sampler_iter is None:
516 self._reset()
--> 517 data = self._next_data()
518 self._num_yielded += 1
519 if self._dataset_kind == _DatasetKind.Iterable and \
~/anaconda3/lib/python3.8/site-packages/torch/utils/data/dataloader.py in _next_data(self)
1180
1181 assert not self._shutdown and self._tasks_outstanding > 0
-> 1182 idx, data = self._get_data()
1183 self._tasks_outstanding -= 1
1184 if self._dataset_kind == _DatasetKind.Iterable:
~/anaconda3/lib/python3.8/site-packages/torch/utils/data/dataloader.py in _get_data(self)
1146 else:
1147 while True:
-> 1148 success, data = self._try_get_data()
1149 if success:
1150 return data
~/anaconda3/lib/python3.8/site-packages/torch/utils/data/dataloader.py in _try_get_data(self, timeout)
984 # (bool: whether successfully get data, any: data if successful else None)
985 try:
--> 986 data = self._data_queue.get(timeout=timeout)
987 return (True, data)
988 except Exception as e:
~/anaconda3/lib/python3.8/multiprocessing/queues.py in get(self, block, timeout)
105 if block:
106 timeout = deadline - time.monotonic()
--> 107 if not self._poll(timeout):
108 raise Empty
109 elif not self._poll():
~/anaconda3/lib/python3.8/multiprocessing/connection.py in poll(self, timeout)
255 self._check_closed()
256 self._check_readable()
--> 257 return self._poll(timeout)
258
259 def __enter__(self):
~/anaconda3/lib/python3.8/multiprocessing/connection.py in _poll(self, timeout)
422
423 def _poll(self, timeout):
--> 424 r = wait([self], timeout)
425 return bool(r)
426
~/anaconda3/lib/python3.8/multiprocessing/connection.py in wait(object_list, timeout)
929
930 while True:
--> 931 ready = selector.select(timeout)
932 if ready:
933 return [key.fileobj for (key, events) in ready]
~/anaconda3/lib/python3.8/selectors.py in select(self, timeout)
413 ready = []
414 try:
--> 415 fd_event_list = self._selector.poll(timeout)
416 except InterruptedError:
417 return ready
KeyboardInterrupt:
4.7. Evaluation Script¶
with torch.no_grad():
model.eval()
correct = total = 0
with torch.no_grad():
for sample in validation_loader:
inputs = sample[0].float().to(device)
inputs = inputs.squeeze(1)
b = inputs.size(0)
t = inputs.size(1)
inputs = inputs.reshape(b, t, -1)
labels = sample[1].long().to(device)
labels = labels.squeeze(1)
output = model(inputs.transpose(1,2))
_, predicted = torch.max(output, -1)
total += labels.size(0)
correct += (predicted == labels).sum().item()
val_acc = 100. * correct / total
print('Valid accuracy: %f %%' % val_acc)
def plot_cm(model, validation_loader):
total = correct = 0
conf_mat = []
with torch.no_grad():
for i,sample in enumerate(validation_loader):
if i % 1 == 0:
inputs = sample[0].float().to(device)
inputs = inputs.squeeze()
inputs = inputs.squeeze(1)
b = inputs.size(0)
t = inputs.size(1)
inputs = inputs.reshape(b,t,-1)
labels = sample[1].long().to(device)
output = model(inputs.transpose(1,2))
_, predicted = torch.max(output, -1)
#print(torch.unique(labels))
#print(torch.unique(predicted))
conf_matrix = metrics.confusion_matrix(predicted.float().cpu(), labels.float().cpu())
conf_mat.append(conf_matrix)
total += labels.size(0)
correct += (predicted.ravel() == labels.ravel()).sum().item()
agg_cm = np.sum(conf_mat,0)
agg_cm = agg_cm/np.sum(agg_cm,0)
classes = np.array(['IMF','BS-crossing','magnetosheath','MP-crossing','magnetosphere'])
con_mat_df = pd.DataFrame(
agg_cm, index = classes,columns = classes)
##plot
figure = plt.figure(figsize=(8, 8))
sns.heatmap(con_mat_df, annot=True,cmap=plt.cm.Blues)
return con_mat_df
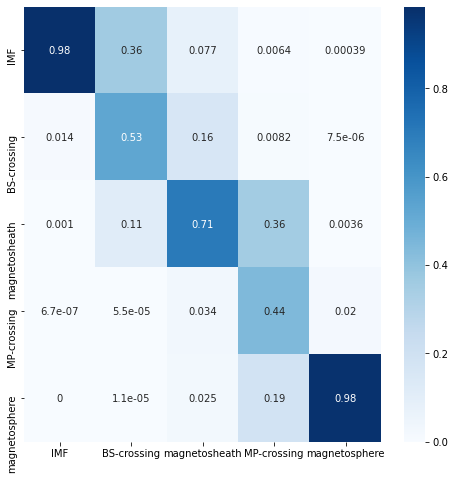
plot_cm(model, validation_loader)
| IMF | BS-crossing | magnetosheath | MP-crossing | magnetosphere | |
|---|---|---|---|---|---|
| IMF | 9.845936e-01 | 0.358994 | 0.077034 | 0.006411 | 0.000386 |
| BS-crossing | 1.439123e-02 | 0.528224 | 0.158833 | 0.008233 | 0.000007 |
| magnetosheath | 1.014515e-03 | 0.112715 | 0.705087 | 0.357031 | 0.003596 |
| MP-crossing | 6.703105e-07 | 0.000055 | 0.033648 | 0.443121 | 0.020106 |
| magnetosphere | 0.000000e+00 | 0.000011 | 0.025398 | 0.185204 | 0.975905 |
def plot_field(df):
fig = go.Figure()
# Plotting components of the magnetic field B_x, B_y, B_z in MSO coordinates
fig.add_trace(go.Scatter(x=df['DATE'], y=df['BX_MSO'], name='B_x'))
fig.add_trace(go.Scatter(x=df['DATE'], y=df['BY_MSO'], name='B_y'))
fig.add_trace(go.Scatter(x=df['DATE'], y=df['BZ_MSO'], name='B_z'))
# Plotting total magnetic field magnitude B along the orbit
fig.add_trace(go.Scatter(x=df['DATE'], y=-df['B_tot'], name='|B|', line_color = 'darkgray'))
fig.add_trace(go.Scatter(x=df['DATE'], y=df['B_tot'], name='|B|', line_color = 'darkgray', showlegend=False))
return fig
def show_plot(df, model, annotation, orbit_no, evaluate = False):
fig = plot_field(df)
df['Label'] = annotation
bowshock = df.loc[df['Label'] == 1]
magnetopause = df.loc[df['Label'] == 3]
for i in range(len(bowshock['DATE'])):
fig.add_trace(go.Scatter(x=[bowshock['DATE'].iloc[i], bowshock['DATE'].iloc[i]],
y=[-450, 450],
mode='lines',
name='Bow Shock',
line_color='black',
showlegend=False
))
for i in range(len(magnetopause['DATE'])):
fig.add_trace(go.Scatter(x=[magnetopause['DATE'].iloc[i], magnetopause['DATE'].iloc[i]],
y=[-450, 450],
mode='lines',
name='Magnetopause',
line_color = 'purple',
showlegend=False
))
print(orbit_no)
if evaluate == True:
fig.update_layout({'title': f'Messenger orbit {orbit_no:04d} [Learned]'})
else:
fig.update_layout({'title': f'Messgener orbit {orbit_no:04d} [Ground Truth]'})
fig.show()
model = CNN(3,6).to(device)
model.load_state_dict(torch.load('model-windowed-cnn.pth'))
<All keys matched successfully>
model_partial = CNN(3,4).to(device)
model_partial.load_state_dict(torch.load('saved_models/model-classification-partial.pth'))
features_list = ['X_MSO', 'Y_MSO', 'Z_MSO','BX_MSO', 'BY_MSO', 'BZ_MSO', 'VX', 'VY', 'VZ', 'EXTREMA', 'COSALPHA', 'RHO_DIPOLE']
def plot_prediction(model,orbit_no, features_list):
orbit_no = int(orbit_no)
total = correct = 0
csv = f'../03-notebooks/labelled_orbits/train/test/df_{orbit_no:d}.csv'
df = pd.read_csv(csv, sep = ',')
label = df.LABEL
df["B_tot"] = (df["BX_MSO"]**2 + df["BY_MSO"]**2 + df["BZ_MSO"]**2)**0.5
orbit = Magnetometer_data(csv, features_list)
annotation = []
for sample in orbit:
coords = torch.tensor(sample['coords']).float().to(device)
coords = torch.unsqueeze(coords,0)
output = model_partial(coords.transpose(1,2))
predicted = output.argmax(-1)
annotation.append(predicted)
show_plot(df, model_partial, annotation, orbit_no, evaluate = True)
show_plot(df, model_partial, label, orbit_no)
plot_prediction(model_partial, 1790, features_list)
<ipython-input-18-edbe06637cdc>:11: UserWarning: To copy construct from a tensor, it is recommended to use sourceTensor.clone().detach() or sourceTensor.clone().detach().requires_grad_(True), rather than torch.tensor(sourceTensor).
coords = torch.tensor(sample['coords']).float().to(device)
1790
1790
